5IZJ - chain B (model A) | Protein kinase cAMP-activated catalytic subunit alpha
Structure information
| PDB: | 5IZJ |
| PubMed: | 27389935 |
| Release date: | 2016-07-20 |
| Resolution: | 1.85 Å |
| Kinase: | PRKACA (PKACa) |
| Family: | PKA |
| Group: | AGC |
| Species: | HUMAN |
| Quality Score: | 8 |
| Missing Residues: | 0 |
| Missing Atoms: | 0 |
| DFG conformation: | in |
| αC-helix conformation: | in |
| Salt bridge KIII.17 and EαC.24: | No (4.5Å) |
| ASP rotation (xDFG.81) : | 336° |
| PHE rotation (xDFG.82) : | 10° |
| Activation loop position: | -4.1Å |
| αC-helix position: | 19.4Å |
| G-rich loop angle: | 66.5° |
| G-rich loop distance: | 20.3Å |
| G-rich loop rotation: | 51.5° |
Other models from this PDB:
2D & 3D views
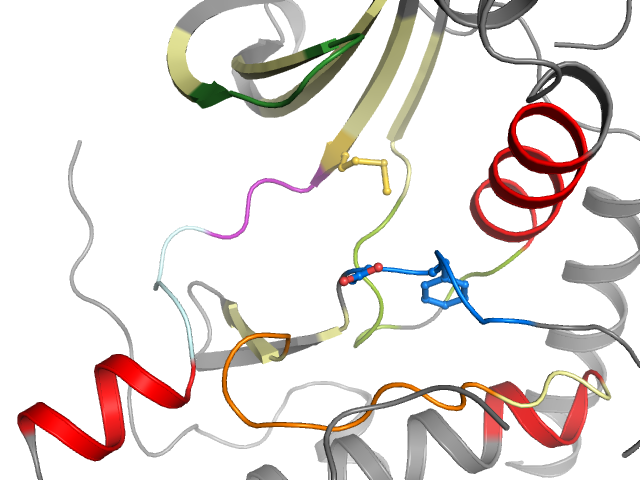
Binding pocket waters
The following waters were found in the defined clusters:
I1
H-bond protein
I3
No H-bonds
I4
H-bond protein
I5
H-bond protein
I9
No H-bonds
Binding pocket sequence
| Uniprot | KTLGTGSFGRVMLYAMKILHTLNEKRILQAVNPFLVKLEFSYMVMEYVPGGEMFSHLRRYLHSLDLIYRDLKPENLLIVTDFGFA |
| Structure: | KTLGTGSFGRVMLYAMKILHTLNEKRILQAVNPFLVKLEFSYMVMEYVPGGEMFSHLRRYLHSLDLIYRDLKPENLLIVTDFGFA |
Modified residues
Residue 197 (not in pocket)
Phosphorylated threonine
Residue 338 (not in pocket)
Phosphorylated serine
Allosteric ligand
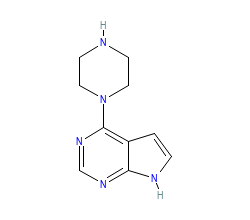
Ligand HET-code: 6J9
Ligand Name: 4-(piperazin-1-yl)-7H-pyrrolo[2,3-d]pyrimidine
Binding affinities
Ligand not found in ChEMBL.