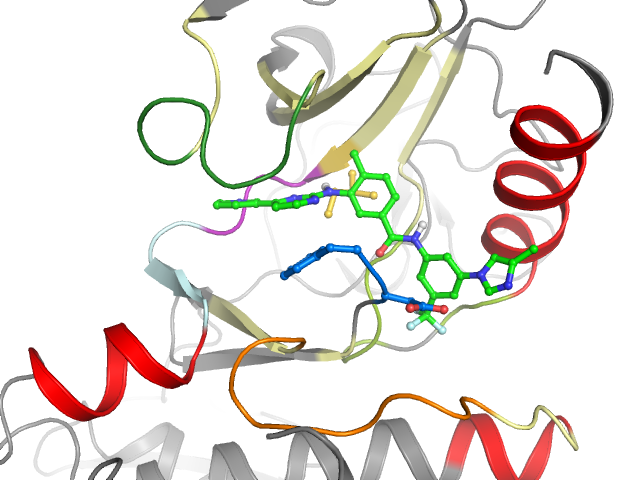
5MO4 - chain A (model B) | ABL proto-oncogene 1, non-receptor tyrosine kinase
Structure information
| PDB: | 5MO4 |
| PubMed: | 28329763 |
| Release date: | 2017-04-05 |
| Resolution: | 2.17 Å |
| Kinase: | ABL1 |
| Family: | Abl |
| Group: | TK |
| Species: | HUMAN |
| Quality Score: | 8 |
| Missing Residues: | 3 |
| Missing Atoms: | 8 |
| DFG conformation: | out |
| αC-helix conformation: | in |
| Salt bridge KIII.17 and EαC.24: | Yes (2.8Å) |
| ASP rotation (xDFG.81) : | 190° |
| PHE rotation (xDFG.82) : | 176° |
| Activation loop position: | 2.1Å |
| αC-helix position: | 18.6Å |
| G-rich loop angle: | 63.1° |
| G-rich loop distance: | 19Å |
| G-rich loop rotation: | 16° |
Other models from this PDB:
2D & 3D views
Binding pocket waters
The following waters were found in the defined clusters:
I1
H-bond protein
I10
H-bond protein
O1
H-bond ligand
Binding pocket sequence
| Uniprot | HKLGGGQYGEVYEVAVKTLEFLKEAAVMKEIKPNLVQLLGVYIITEFMTYGNLLDYLREYLEKKNFIHRDLAARNCLVVADFGLS |
| Structure: | HKLGGGQYGEVYEVAVKTLEFLKEAAVMKEIKPNLVQLLGVYIIIEFMTYGNLLDYLREYLEKKNFIHRNLAARNCLVVADF___ |
Modified residues
No modified residues identified.
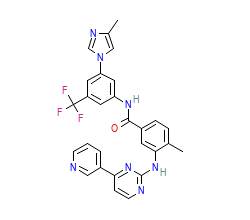
Orthosteric ligand
- Download image
- LABELS
- KLIFS residue #
- Amino Acid
- None
- COLORS
- Interaction types
- KLIFS (all res.)
- KLIFS (interacting res.)
- None
- OTHER
- Show/hide non-interacting res.
- En/disable resizing interacting res.
This ligand targets the following (sub)pockets:
| Main pockets | |
|---|---|
| Front | |
| Gate | |
| Back | |
| Subpockets | |
|---|---|
| FP-I | |
| FP-II | |
| BP-I-A | |
| BP-I-B | |
| BP-II-in | |
| BP-II-A-in | |
| BP-II-B-in | |
| BP-II-out | |
| BP-II-B | |
| BP-III | |
| BP-IV | |
| BP-V | |
Kinase-ligand interactions
■ Hydrophobic ♦ Aromatic face-to-face ♦ Aromatic face-to-edge ▲ H-bond donor ▲ H-bond acceptor ● Ionic positive ● Ionic negative
| I | g.l | II | III | αC | |||||||||||||||
| 1 H 265 | 2 K 266 | 3 L 267 | 4 G 268 | 5 G 269 | 6 G 270 | 7 Q 271 | 8 Y 272 | 9 G 273 | 10 E 274 | 11 V 275 | 12 Y 276 | 13 E 277 | 14 V 287 | 15 A 288 | 16 V 289 | 17 K 290 | 18 T 291 | 19 L 292 | 20 E 301 |
| ■ | ■♦ | ■ | ■ | ■ | ■ | ||||||||||||||
| αC | b.l | IV | |||||||||||||||||
| 21 F 302 | 22 L 303 | 23 K 304 | 24 E 305 | 25 A 306 | 26 A 307 | 27 V 308 | 28 M 309 | 29 K 310 | 30 E 311 | 31 I 312 | 32 K 313 | 33 P 315 | 34 N 316 | 35 L 317 | 36 V 318 | 37 Q 319 | 38 L 320 | 39 L 321 | 40 G 322 |
| ■ | ■▲ | ■ | ■ | ■ | ■ | ■ | |||||||||||||
| IV | V | GK | hinge | linker | αD | αE | |||||||||||||
| 41 V 323 | 42 Y 331 | 43 I 332 | 44 I 333 | 45 I 334 | 46 E 335 | 47 F 336 | 48 M 337 | 49 T 338 | 50 Y 339 | 51 G 340 | 52 N 341 | 53 L 342 | 54 L 343 | 55 D 344 | 56 Y 345 | 57 L 346 | 58 R 347 | 59 E 348 | 60 Y 372 |
| ■ | ■ | ■♦ | ■▲ | ■ | |||||||||||||||
| αE | VI | c.l | VII | VIII | x | ||||||||||||||
| 61 L 373 | 62 E 374 | 63 K 375 | 64 K 376 | 65 N 377 | 66 F 378 | 67 I 379 | 68 H 380 | 69 R 381 | 70 N 382 | 71 L 383 | 72 A 384 | 73 A 385 | 74 R 386 | 75 N 387 | 76 C 388 | 77 L 389 | 78 V 390 | 79 V 398 | 80 A 399 |
| ■ | ■♦ | ■ | ■ | ■ | ■ | ||||||||||||||
| DFG | a.l | ||||||||||||||||||
| 81 D 400 | 82 F 401 | 83 _ _ | 84 _ _ | 85 _ _ | |||||||||||||||
| ■▲ | ■♦ | ||||||||||||||||||
Binding affinities
ChEMBL ID:CHEMBL255863Bioaffinities: 136 records for 59 kinase(s)
| Species | Kinase (ChEMBL naming) | Median | Min | Max | Type | Records |
|---|---|---|---|---|---|---|
| Homo sapiens | Breakpoint cluster region protein | 6.2 | 6.2 | 6.2 | pKd | 2 |
| Plasmodium falciparum | Calcium-dependent protein kinase 1 | 6.1 | 6.1 | 6.1 | pKd | 1 |
| Homo sapiens | c-Jun N-terminal kinase 1 | 6.4 | 6.4 | 6.4 | pKd | 1 |
| Homo sapiens | c-Jun N-terminal kinase 2 | 5.2 | 5.2 | 5.2 | pKd | 1 |
| Homo sapiens | c-Jun N-terminal kinase 3 | 5.7 | 5.7 | 5.7 | pKd | 1 |
| Homo sapiens | Discoidin domain-containing receptor 2 | 7.8 | 7 | 8.7 | pIC50 | 5 |
| Homo sapiens | Discoidin domain-containing receptor 2 | 7.5 | 6.5 | 8.2 | pKd | 5 |
| Homo sapiens | Dual specificity mitogen-activated protein kinase kinase 5 | 6.7 | 6.7 | 6.7 | pKd | 1 |
| Homo sapiens | Dual specificity protein kinase CLK4 | 5.2 | 5.2 | 5.2 | pKd | 1 |
| Homo sapiens | Dual specificty protein kinase CLK1 | 5.7 | 5.7 | 5.7 | pKd | 2 |
| Homo sapiens | Ephrin type-A receptor 1 | 6.2 | 6.2 | 6.2 | pKd | 1 |
| Homo sapiens | Ephrin type-A receptor 2 | 6.1 | 6.1 | 6.6 | pKd | 2 |
| Homo sapiens | Ephrin type-A receptor 3 | 7 | 7 | 7 | pKd | 1 |
| Homo sapiens | Ephrin type-A receptor 4 | 6.5 | 6.5 | 6.5 | pKd | 1 |
| Homo sapiens | Ephrin type-A receptor 5 | 5.7 | 5.7 | 5.7 | pKd | 1 |
| Homo sapiens | Ephrin type-A receptor 6 | 6.2 | 6.2 | 6.2 | pKd | 1 |
| Homo sapiens | Ephrin type-A receptor 8 | 7.4 | 7.4 | 7.4 | pKd | 1 |
| Homo sapiens | Ephrin type-B receptor 1 | 5.9 | 5.9 | 5.9 | pKd | 1 |
| Homo sapiens | Ephrin type-B receptor 2 | 6.2 | 6.2 | 6.2 | pKd | 1 |
| Homo sapiens | Ephrin type-B receptor 3 | 6 | 6 | 6 | pKd | 1 |
| Homo sapiens | Ephrin type-B receptor 4 | 6.1 | 6.1 | 6.1 | pKd | 1 |
| Homo sapiens | Ephrin type-B receptor 6 | 6.3 | 6.3 | 6.3 | pKd | 1 |
| Homo sapiens | Epithelial discoidin domain-containing receptor 1 | 7.5 | 7.4 | 8.4 | pIC50 | 3 |
| Homo sapiens | Epithelial discoidin domain-containing receptor 1 | 6.9 | 6.9 | 9 | pKd | 2 |
| Homo sapiens | Macrophage colony stimulating factor receptor | 6.2 | 6.2 | 6.2 | pIC50 | 1 |
| Homo sapiens | Macrophage colony stimulating factor receptor | 7.4 | 7.4 | 7.4 | pKd | 1 |
| Homo sapiens | MAP kinase p38 alpha | 6.3 | 6 | 6.3 | pKd | 3 |
| Homo sapiens | MAP kinase p38 beta | 5.4 | 5.4 | 7.4 | pKd | 2 |
| Homo sapiens | Mitogen-activated protein kinase kinase kinase kinase 1 | 6.1 | 6.1 | 6.1 | pKd | 1 |
| Homo sapiens | Mixed lineage kinase 7 | 6 | 6 | 8 | pKd | 2 |
| Homo sapiens | Neurotrophic tyrosine kinase receptor type 2 | 6.3 | 6.3 | 6.3 | pKd | 1 |
| Homo sapiens | NT-3 growth factor receptor | 6.2 | 6.2 | 6.2 | pKd | 1 |
| Homo sapiens | Phosphatidylinositol-5-phosphate 4-kinase type-2 gamma | 5.1 | 5.1 | 5.1 | pKd | 1 |
| Homo sapiens | Phosphorylase kinase gamma subunit 2 | 6.5 | 6.5 | 6.5 | pKd | 1 |
| Homo sapiens | Platelet-derived growth factor receptor alpha | 7.2 | 7.2 | 7.2 | pIC50 | 1 |
| Homo sapiens | Platelet-derived growth factor receptor alpha | 6.8 | 6.8 | 7.1 | pKd | 2 |
| Homo sapiens | Platelet-derived growth factor receptor beta | 7.2 | 7.2 | 7.6 | pIC50 | 3 |
| Homo sapiens | Platelet-derived growth factor receptor beta | 7.1 | 7.1 | 7.8 | pKd | 2 |
| Homo sapiens | Receptor-interacting serine/threonine-protein kinase 3 | 5.9 | 5.9 | 5.9 | pKd | 1 |
| Homo sapiens | Serine/threonine-protein kinase 10 | 5.2 | 5.2 | 5.2 | pKd | 1 |
| Homo sapiens | Serine/threonine-protein kinase B-raf | 5.8 | 5.8 | 6.2 | pKd | 2 |
| Homo sapiens | Serine/threonine-protein kinase ILK-1 | 7.7 | 7.7 | 7.7 | pKd | 1 |
| Homo sapiens | Serine/threonine-protein kinase MRCK beta | 6 | 6 | 6 | pKd | 1 |
| Homo sapiens | Serine/threonine-protein kinase RAF | 6 | 6 | 6 | pIC50 | 1 |
| Homo sapiens | Serine/threonine-protein kinase RAF | 5.4 | 5.4 | 5.4 | pKd | 1 |
| Homo sapiens | Serine/threonine-protein kinase TAO1 | 5.3 | 5.3 | 5.3 | pKd | 1 |
| Homo sapiens | Serine/threonine-protein kinase TAO3 | 5.8 | 5.8 | 5.8 | pKd | 1 |
| Homo sapiens | Serine/threonine-protein kinase TNNI3K | 6.4 | 6.4 | 6.4 | pKd | 1 |
| Homo sapiens | Stem cell growth factor receptor | 6.8 | 6.7 | 7.8 | pIC50 | 4 |
| Homo sapiens | Stem cell growth factor receptor | 7.3 | 6.1 | 7.8 | pKd | 9 |
| Homo sapiens | Tyrosine-protein kinase ABL | 7.6 | 6.2 | 8.9 | pIC50 | 12 |
| Homo sapiens | Tyrosine-protein kinase ABL | 7.9 | 6.2 | 8.4 | pKd | 17 |
| Homo sapiens | Tyrosine-protein kinase ABL2 | 6.5 | 6.5 | 7.6 | pKd | 2 |
| Homo sapiens | Tyrosine-protein kinase BLK | 6.3 | 6.3 | 6.3 | pKd | 1 |
| Homo sapiens | Tyrosine-protein kinase CSK | 5.6 | 5.6 | 5.6 | pKd | 1 |
| Homo sapiens | Tyrosine-protein kinase FGR | 6.5 | 6.5 | 6.5 | pKd | 1 |
| Homo sapiens | Tyrosine-protein kinase FRK | 6.3 | 6.3 | 7.1 | pKd | 2 |
| Homo sapiens | Tyrosine-protein kinase FYN | 5.8 | 5.8 | 5.8 | pKd | 1 |
| Homo sapiens | Tyrosine-protein kinase HCK | 6.4 | 6.4 | 6.4 | pKd | 1 |
| Homo sapiens | Tyrosine-protein kinase LCK | 7 | 7 | 7 | pIC50 | 1 |
| Homo sapiens | Tyrosine-protein kinase LCK | 7.3 | 7.3 | 7.3 | pKd | 1 |
| Homo sapiens | Tyrosine-protein kinase Lyn | 5.9 | 5.9 | 5.9 | pIC50 | 1 |
| Homo sapiens | Tyrosine-protein kinase Lyn | 5.8 | 5.8 | 7 | pKd | 2 |
| Homo sapiens | Tyrosine-protein kinase receptor FLT3 | 5 | 5 | 5 | pIC50 | 1 |
| Homo sapiens | Tyrosine-protein kinase receptor FLT3 | 4.4 | 4.4 | 4.4 | pKd | 1 |
| Homo sapiens | Tyrosine-protein kinase receptor RET | 6 | 6 | 6.1 | pKd | 2 |
| Homo sapiens | Tyrosine-protein kinase receptor Tie-1 | 6 | 6 | 6 | pKd | 1 |
| Homo sapiens | Tyrosine-protein kinase SRC | 5.7 | 5.7 | 5.7 | pIC50 | 1 |
| Homo sapiens | Tyrosine-protein kinase SRC | 5.7 | 5.7 | 5.7 | pKd | 1 |
| Homo sapiens | Tyrosine-protein kinase TIE-2 | 6 | 6 | 6 | pKd | 1 |
| Homo sapiens | Tyrosine-protein kinase YES | 6 | 6 | 6 | pKd | 1 |
Allosteric ligand
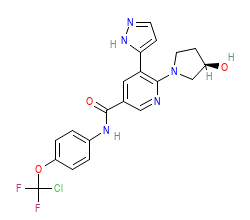
Ligand HET-code: AY7
Ligand Name: asciminib
Binding affinities
Ligand not found in ChEMBL.